Our tool enables more accurate identification of CRISPR targets on non-coding genes for the CRISPRko and CRISPRi mechanisms.
Currently existing sgRNA design tools are developed for coding genes, and our tool CRISPRlnc compensates for the lack of specialized tools for non-coding genes.
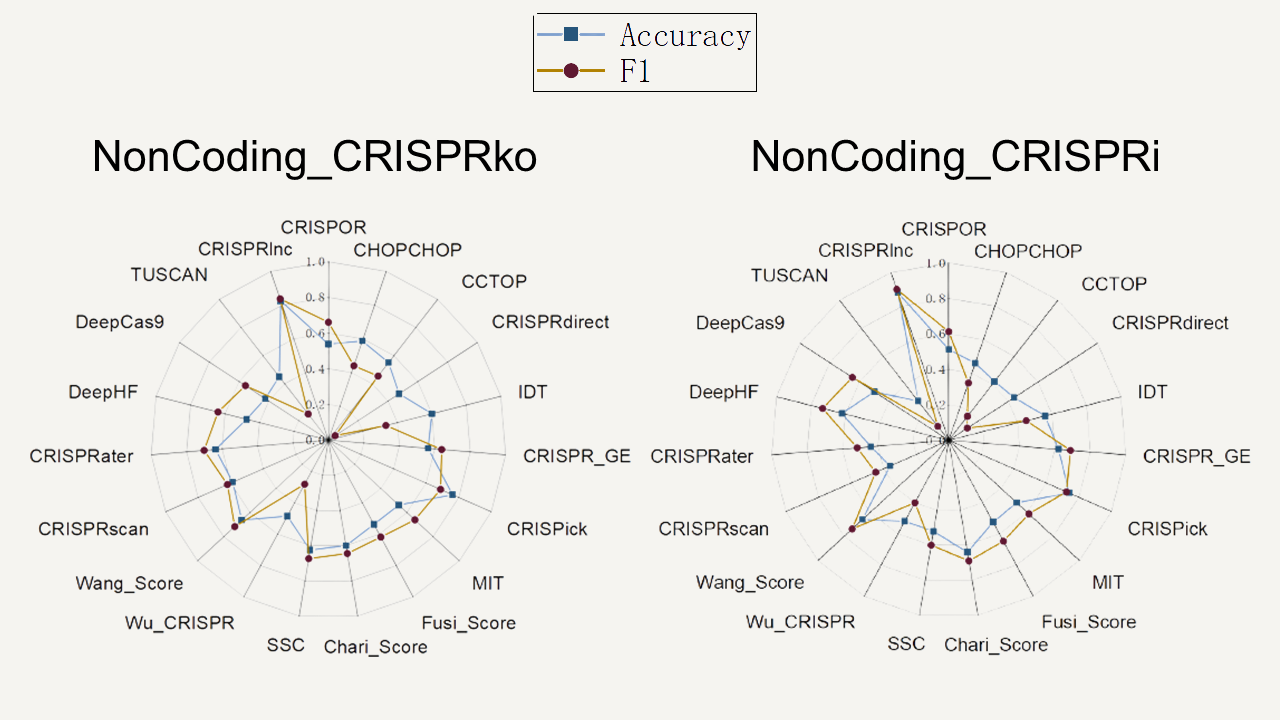
To validate the predictive performance of CRISPRlnc, we compared its performance with other tools (attached below is a list of software participating in the performance evaluation) using independent test datasets, and the results showed that our tool could more accurately identify CRISPR targets on non-coding genes in CRISPRko and CRISPRi mechanisms (as shown in the left figures).